|
RESOURCES AND INSTRUMENTATION >
INSTRUMENTATION

Sequencing: Illumina
 |
Illumina NextSeq 500
This system can configured to meet researchers' individual needs.
- Reads up to 150 bp long
- Single Read or Paired End
- Two flow cell sizes:
- High - up to 400 million clusters
- Mid - up to 130 million clusters
- Indexing/barcoding supported
For more information on the HiSeq, see our Illumina techniques page, or visit http://www.illumina.com.
Before beginnig a project, please contact Christian Daly (cdaly@cgr.harvard.edu) to discuss your experimental set-up. For pricing, please see the price list. |
 |
Illumina HiSeq 2500
This system can can be configured to meet researchers' individual needs.
- Reads up to 150 bp long
- Single Read or Paired End
- Two flow cell sizes:
- "High Output" v4 - 8 lanes, up to 250 million clusters each
- Rapid Run - 2 lanes, up to 150 million clusters each
- Indexing/barcoding supported
For more information on the HiSeq, see our Illumina techniques page, or visit http://www.illumina.com.
Before beginnig a project, please contact Christian Daly (cdaly@cgr.harvard.edu) to discuss your experimental set-up. For pricing, please see the price list. |
 |
Illumina HiSeq 2000
This system can generate up to 200 million clusters per sample and is usually used for short reads using standard "high output" v3 chemistry. Indexing/barcoding is supported.
For more information on the HiSeq, see our Illumina techniques page, or visit http://www.illumina.com.
Before beginnig a project, please contact Christian Daly (cdaly@cgr.harvard.edu) to discuss your experimental set-up. For pricing, please see the price list. |
 |
IntegenX Apollo 324
This instrument automates next-generation sequencing library preparation using bead technology.
- Reagent kits are available for DNA sequencing, RNA-Seq or ChIP-seq.
- Compatible with indexing/multiplexing.
- Prepare 1-48 samples at a time.
For more information on the Apollo 324, see our Illumina techniques page or visit http://integenx.com/products/apollo-324/.
To learn about using this instrument, please contact Jennifer Couget (jcouget@cgr.harvard.edu, 617-495-9402). |
Ancillary Equipment for Sequencing Projects
The core offers instruments for sample shearing, size selection, library QC and quantification.
|
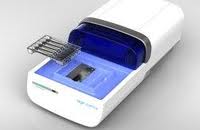 |
Pippin Prep
This instrument automates nucleic acid size selection using convenient gel cassettes.
- Choose broad or tight size range cuts.
- Choose the % gel that will allow for selection of your desired size.
- Prepare 4 samples at a time along with a reference.
For more information on the Pippin Prep, see our Illumina techniques page or visit http://www.sagescience.com/.
To learn about using this instrument, please contact Jennifer Couget (jcouget@cgr.harvard.edu, 617-495-9402) |
 |
Life Tech E-gel system
Together the E-gel iBase and Safe Imager automate nucleic acid electrophoresis and size selection using pre-cast gels. Features include fast run times, programmable control, and real time monitoring of DNA migration. Researchers must supply their own gels for use on the system.
For more information on the Pippin Prep, see our Illumina techniques page or visit http://www.lifetechnologies.com.
To learn about using this instrument, please contact Jennifer Couget (jcouget@cgr.harvard.edu, 617-495-9402) |
| |
|